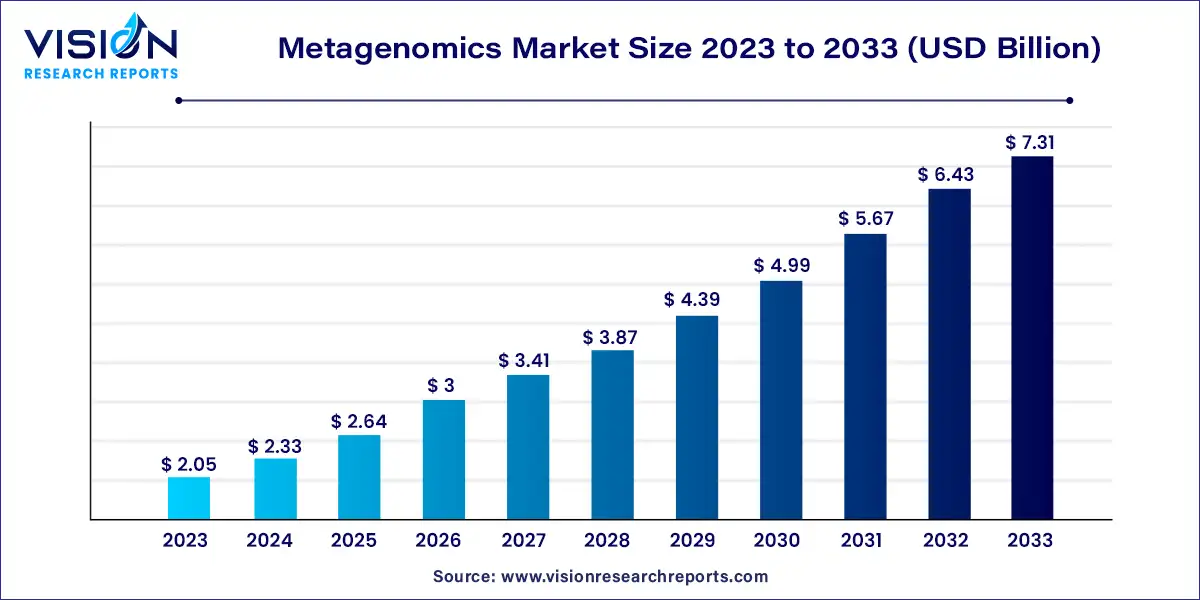
The global metagenomics market size was estimated at around USD 2.05 billion in 2023 and it is projected to hit around USD 7.31 billion by 2033, growing at a CAGR of 13.55% from 2024 to 2033.
Key Pointers
- North America dominated the market in 2023, holding a 41% share.
- Asia Pacific is projected to experience the most rapid growth in the market from 2024 to 2033.
- Kits and reagents were the most lucrative product segment in 2023, accounting for 62% of market revenue.
- Sequencing and data analytics services are anticipated to expand at the fastest rate during the forecast period.
- Shotgun sequencing was the dominant technology in 2023, capturing 35% of the market.
- 16S sequencing is expected to see substantial growth in the coming years.
- Sequencing was the primary workflow in 2023, comprising 54% of the market.
- Data analysis is projected to have the highest growth rate among workflows during the forecast period.
- Environmental applications contributed the most to market revenue in 2023, with a 26% share.
Get Sample@ https://www.visionresearchreports.com/report/sample/41293
What is Metagenomics
Metagenomics is the study of genetic material recovered directly from environmental samples, rather than from cultured organisms. This field of genomics allows researchers to analyze the collective genomes of microorganisms in a specific environment, which could include soil, water, the human gut, or any other ecosystem.
Key Aspects of Metagenomics:
- Environmental Samples: Metagenomics focuses on studying DNA extracted from environmental samples containing diverse microbial communities, such as bacteria, viruses, fungi, and archaea.
- Unculturable Microbes: Traditional microbiology relies on culturing organisms in a lab, but many microbes cannot be easily cultured. Metagenomics overcomes this limitation by allowing scientists to study these unculturable microbes directly from their natural environments.
- High-Throughput Sequencing: The advent of next-generation sequencing (NGS) technologies has significantly advanced metagenomics, enabling the rapid and cost-effective sequencing of complex microbial communities.
- Applications: Metagenomics has broad applications, including understanding microbial diversity, discovering new genes and enzymes, studying the microbiome’s role in human health and disease, environmental monitoring, bioremediation, and even forensic analysis.
- Data Analysis: Metagenomics involves sophisticated bioinformatics tools to analyze vast amounts of sequencing data, identifying species, gene functions, and interactions within microbial communities.
Advances in Metagenomics and Its Application in Environmental Microorganisms
Advances in Metagenomics
Metagenomics has seen significant advancements over the past decade, largely driven by innovations in sequencing technologies, bioinformatics tools, and computational power. These advances have enabled deeper insights into the complex and diverse microbial communities found in various environments.
- Next-Generation Sequencing (NGS): The introduction of high-throughput sequencing technologies, such as Illumina and PacBio, has revolutionized metagenomics by allowing researchers to rapidly sequence entire microbial communities. This has enabled the identification of millions of microbial species and their genetic functions without the need for culturing.
- Shotgun Metagenomics: This approach involves randomly sequencing all the DNA in a sample, providing a comprehensive overview of the microbial community’s genetic composition. Shotgun metagenomics allows for the discovery of novel genes, metabolic pathways, and microbial interactions.
- Long-Read Sequencing: Technologies like PacBio and Oxford Nanopore provide longer DNA reads, which help in assembling more complete genomes from metagenomic data. This is crucial for identifying and characterizing species with high accuracy and resolving complex microbial communities.
- Single-Cell Metagenomics: This technique allows the study of genomes from individual cells within a microbial community, enabling the characterization of rare or unculturable microbes. Single-cell metagenomics is particularly useful for studying microbial diversity and evolution.
- Bioinformatics and Data Analysis: The development of advanced bioinformatics tools and pipelines, such as MetaPhlAn, QIIME, and MEGAHIT, has enhanced the analysis of metagenomic data. These tools allow researchers to accurately classify species, predict gene functions, and reconstruct metabolic pathways, providing deeper insights into microbial ecosystems.
Applications in Environmental Microorganisms
Metagenomics has transformative applications in studying environmental microorganisms, offering insights into microbial diversity, ecosystem functioning, and environmental sustainability.
- Microbial Diversity and Ecology: Metagenomics enables the exploration of microbial diversity in various environments, such as soil, oceans, and extreme habitats like hydrothermal vents. By analyzing the metagenomes of these environments, scientists can uncover the roles of different microbes in nutrient cycling, organic matter decomposition, and ecosystem stability.
- Biogeochemical Cycles: Metagenomics has been instrumental in understanding how microorganisms contribute to global biogeochemical cycles, such as carbon, nitrogen, and sulfur cycles. For instance, metagenomic studies have identified key microbial players involved in methane production in wetlands or nitrogen fixation in agricultural soils.
- Bioremediation: Metagenomics is used to identify and monitor microbial communities involved in the degradation of pollutants, such as hydrocarbons, heavy metals, and plastics. By understanding the genetic potential of these communities, scientists can develop strategies to enhance bioremediation processes in contaminated environments.
- Climate Change Research: The study of environmental microorganisms through metagenomics helps in assessing the impact of climate change on microbial communities and their functions. For example, metagenomic analyses of polar regions and permafrost soils reveal how warming temperatures influence microbial activity and greenhouse gas emissions.
- Agricultural and Soil Health: Metagenomics is applied to study soil microbiomes, helping to improve soil health and crop productivity. By analyzing soil microbial communities, researchers can identify beneficial microbes that promote plant growth or suppress plant pathogens, leading to more sustainable agricultural practices.
- Marine and Aquatic Environments: In marine environments, metagenomics has provided insights into the microbial communities that drive primary production, nutrient cycling, and the degradation of organic matter. These studies are crucial for understanding the health of marine ecosystems and their response to environmental changes.
Metagenomics Market Top Companies
Read More@ https://www.heathcareinsights.com/cell-viability-assays-market/
Metagenomics Market Segmentations:
By Product
- Kits & Reagents
- Sequencing & Data Analytics Services
- Software
By Technology
- Shotgun Sequencing
- 16S Sequencing
- Whole Genome Sequencing
- Others
By Workflow
- Pre-sequencing
- Sequencing
- Data Analysis
By Application
- Environmental
- Clinical Diagnostics
- Drug Discovery
- Biotechnology
- Food & nutrition
- Others
By Region
- North America
- Europe
- Asia Pacific
- Latin America
- Middle East & Africa
Buy this Premium Research Report@ https://www.visionresearchreports.com/report/checkout/41293
You can place an order or ask any questions, please feel free to contact sales@visionresearchreports.com| +1 650-460-3308
